Excitability, synaptic transmission, and microcircuits
High-resolution voltage-imaging tools provide a unique window into brain function. By resolving subthreshold events in vivo, voltage imaging can probe the synaptic inputs to a neural circuit. In combination with optogenetic activation or silencing, voltage imaging can resolve the separate roles of synaptic excitation and inhibition. We are using these tools for detailed studies of cortical and hippocampal microcircuits in mice, and of spinal microcircuits in zebrafish.
In vitro voltage imaging provides high-throughput measures of intrinsic excitability of neurons in culture or in acute brain slices, and of synaptic transmission between genetically defined neuronal sub-types. These assays can probe in detail the physiological roles of genetic or pharmacological perturbations and are a powerful tool for disease modeling and drug discovery.
Studies in vivo
- L.Z. Fan, S. Kheifets, U.L. Boehm, H. Wu, K.D. Piatkevich, M.E. Xie, V. Parot, Y. Ha, K.E. Evans, E.S. Boyden, A.E. Takesian, A.E. Cohen, All-Optical Electrophysiology Reveals the Role of Lateral Inhibition in Sensory Processing in Cortical Layer 1, Cell, 180, 521-535, 2020.
- Y. Adam, J.J. Kim, S. Lou, Y. Zhao, M.E. Xie, D. Brinks, H. Wu, M.A. Mostajo-Radji, S. Kheifets, V. Parot, S. Chettih, K.J. Williams, B. Gmeiner, S.L. Farhi, L. Madisen, E.K. Buchanan, I. Kinsella, D. Zhou, L. Paninski, C.D. Harvey, H. Zeng, P. Arlotta, R.E. Campbell, A.E. Cohen, Voltage imaging and optogenetics reveal behaviour-dependent changes in hippocampal dynamics , Nature, 569, 413-417, 2019.
- S. Lou, Y. Adam, E.N. Weinstein, E.Williams, K. Williams, V. Parot, N. Kavokine, S. Liberles, L. Madisen, H. Zeng, A.E. Cohen, Genetically targeted all-optical electrophysiology with a transgenic Cre-dependent Optopatch mouse, The Journal of Neuroscience, 43, 11059-11073, 2016.
Studies in vitro
- L.Z. Fan, R. Nehme, Y. Adam, E.S. Jung, H. Wu, K. Eggan, D.B. Arnold, A.E. Cohen, All-optical synaptic electrophysiology probes mechanism of ketamine-induced disinhibition, Nature Methods, 15, 823-831, 2018. Supplementary Information.
- E. Kiskinis, J.M. Kralj, P. Zou, E.N. Weinstein, H. Zhang, K. Tisoras, O. Wiskow, J.A. Ortega, K. Eggan, A.E. Cohen, All-optical electrophysiology for high-throughput functional characterization of a human iPSC-derived motor neuron model of ALS, Stem Cell Reports, 10, 1-14, 2018.
- K. Ogden, W. Chen, S.A. Swanger, M.J. McDaniel, L.Z.Fan, C. Hu, A. Tankovic, H. Kusumoto, G.J. Kosobucki, A.J. Schulien, Z. Su, J. Pecha, S. Bhattacharya, S. Petrovski, A.E. Cohen, E. Aizenman, S.F. Traynelis, H. Yuan, Molecular mechanism of disease-associated mutations in the pre-M1 helix of NMDA receptors and potential rescue pharmacology, PLoS Genetics, 13, e1006536, 2017.
Voltage imaging tool development
Microbial rhodopsin proteins are found in aquatic organisms throughout the world and in all kingdoms of life. In the wild, these proteins transduce sunlight into changes in membrane potential, which their host organisms use for phototaxis and solar energy capture. We engineered microbial rhodopsins to run in reverse: to transduce changes in membrane potential into an optical signal. These voltage-indicating proteins provide a fast and sensitive tool for monitoring electrical dynamics in cells.
To be used effectively, reporter proteins must be imaged via high-speed and high-sensitivity microscopes. The experiments produce torrents of data, which must be stored and analyzed. Our technology development efforts span from protein engineering to software development.
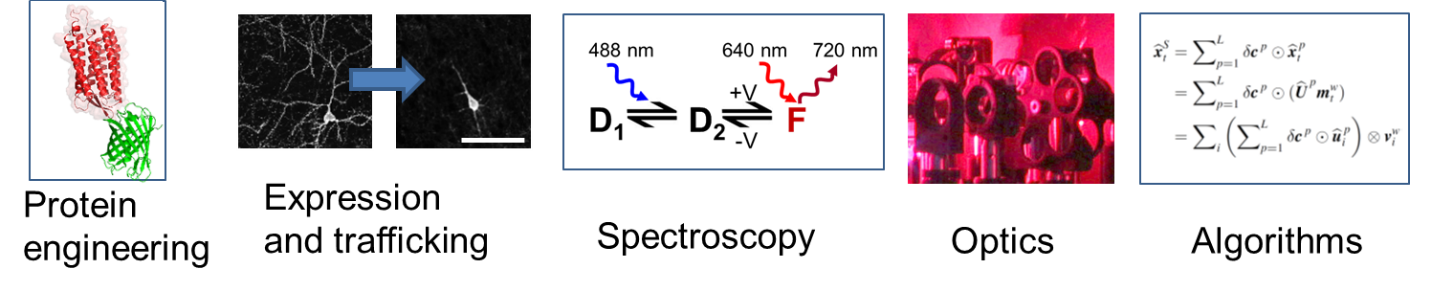
Opsin spectroscopy and engineering
- D. Brinks, A.J. Klein, A.E. Cohen, Two-photon lifetime imaging of voltage indicating proteins as a probe of absolute membrane voltage, Biophysical Journal, 109, 914-921, 2015.
- D.R. Hochbaum*, Y. Zhao*, S.L. Farhi, N. Klapoetke, C.A. Werley, V. Kapoor, P. Zou, J.M. Kralj, D. Maclaurin, N. Smedemark-Margulies, J.L. Saulnier, G.L. Boulting, C. Straub, Y.K. Cho, M. Melkonian, G.K-S. Wong, D.J. Harrison, V.N. Murthy, B.L. Sebatini, E.S. Boyden, R.E. Campbell, A.E. Cohen (*Co-first authors), All-optical electrophysiology in mammalian neurons using engineered microbial rhodopsins, Nature Methods, 11, 825-833, 2014.
- P. Zou*, Y. Zhao*, A.D. Doughlass, D.R. Hochbaum, D. Brinks, C.A. Werley, D.J. Harrison, R.E. Campbell, A.E. Cohen (*Co-first authors), Bright and fast multicoloured voltage reporters via electrochromic FRET, Nature Communications, 5, 2014.
- V. Venkatachalam, A.E. Cohen, Imaging GFP-based reporters in neurons with multiwavelength optogenetic control, Biophys. J., 107, 1554-1563, 2014.
- J. Hou, V. Venkatachalam, A.E. Cohen, Temporal dynamics of microbial rhodopsin fluorescence reports absolute membrane voltage, Biophysical Journal, 106.3, 639-648, 2014.
- J. Park , C.A. Werley, V. Venkatachalam, J.M. Kralj, S.D. Dib-Hajj, S.G. Waxman, A.E. Cohen, Screening fluorescent voltage indicators with spontaneously spiking HEK cells, PLoS ONE, 8, e85221, 2013.
- D. Maclaurin*, V. Venkatachalam*, H. Lee, A.E. Cohen, Mechanism of voltage-sensitive fluorescence in a microbial rhodopsin, PNAS, 110, 5939-5944, 2013.
- V. Venkatachalam, D. Brinks, D. Maclaurin, D. Hochbaum, J.M. Kralj, A.E. Cohen, Flash memory: photochemical imprinting of neuronal action potentials onto a microbial rhodopsin, J. Am. Chem. Soci., 136, 2529-2537 2013.
- H. Bayraktar, A. P. Fields, J. M. Kralj, J. L. Spudich, K. J. Rothschild, A. E. Cohen, Ultrasensitive measurements of microbial rhodopsin photocycles using photochromic FRET, Photochem. & Photobiol. 88, 90-97, 2012.

Microscopes and algorithms
- S.L. Farhi, V.J. Parot, A. Grama, M. Yamagata, A.S. Abdelfattah, Y. Adam, S. Lou, J. J. Kim, R.E. Campbell, D.D. Cox, A.E. Cohen, Wide-area all-optical neurophysiology in acute brain slices, The Journal of Neuroscience, 10, 0168-19, 2019.
- V. J. Parot, C. Sing-Long, Y. Adam, U. L. Bohm, L. Z. Fan, S. L. Farhi, A.E. Cohen, Compressed Hadamard microscopy for high-speed optically sectioned neuronal activity recordings, J. Phys. D: Appl. Phys., 52, 144001, 2019.
- C.A.Werley, M.P.Chien, A.E.Cohen, Ultrawidefield microscope for high-speed fluorescence imaging and targeted optogenetic stiumation, Biomedical Optics Express, 8, 5794-5813, 2017.
Voltage imaging throughout life
Every membrane-enclosed structure can, in principle, support a voltage difference between the inside and outside. We have developed tools to combine optical perturbation and imaging of voltage in systems across scales of biological organization, including:
- Bacteria
- Human stem-cell-derived cardiomyocytes
- Engineered excitable cell lines
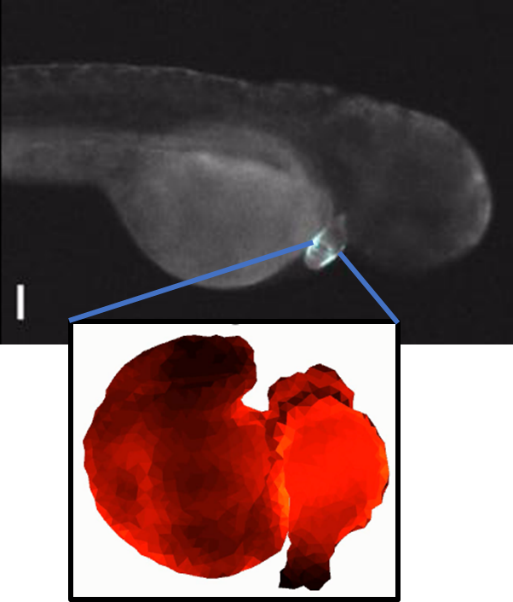
- Zebrafish heart in vivo
Many of the approximations and intuitions developed for neuroscience do not apply in the context of systems with vastly different sizes, ionic compositions, or charge carriers. We are interested in exploring bioelectrical signaling in unconventional regimes.
References
- H.Zhang, E.Reichert, A.E. Cohen, Optical electrophysiology for probing function and pharmacology of voltage-gated ion channels, eLife, 5, e15202, 2016.
- G. T. Dempsey, K.W. Chaudhary, N. Atwater, C. Nguyen, B.S. Brown, J.D. McNeish, A.E. Cohen, J.M. Kralj, Cardiotoxicity screening with simultaneous optogenetic pacing, voltage imaging and calcium imaging, Journal of Pharmacological and Toxicological Methods, 2016.
- J. Hou, J.M. Kralj, A.D. Douglass, F. Engert, A.E. Cohen, Simultaneous mapping of membrane voltage and calcium in zebrafish heart in vivo reveals chamber-specific development transitions in ionic currents, Front. Physiol., 5, 344, 2014. Supporting videos.
- J. M. Kralj, D. R. Hochbaum, A. D. Douglass, A. E. Cohen, Electrical spiking in Escherichia coli probed with a fluorescent voltage-indicating protein, Science, 333, 345-348, 2011. Supporting Online Text and Videos.
Synthetic electrophysiology and bioelectrical patterning
The Hodgkin-Huxley equations of electrophysiology have the same mathematical structure as the Turing reaction-diffusion equation which is often used to describe biological pattern formation during embryonic development. We are studying the interactions of geometry, topology and electrophysiology. How does the geometry of a tissue affect its excitability properties? Can bioelectrical systems spontaneously break spatial symmetry to form steady-state patterns of membrane voltage? What roles does bioelectrical signaling play in embryonic development?
These studies combine cell lines engineered to express defined sets of ion channels and physiological experiments in developing embryos.
References
- H.M. McNamara, R. Salegame, Z. Al Tanoury, H. Xu, S. Begum, G. Ortiz, O. Pourquie, A.E. Cohen Bioelectrical domain walls in homogeneous tissues, Nature Physics, https://doi.org/10.1038/s41567-019-0765-4, 2020.
- H.M. McNamara, S. Dodson, Y. Huang, E.W. Miller, B. Sandstede, A.E. Cohen, Geometry-dependent arrhythmias in electrically excitable tissues, Cell Systems, 7, 359-370, 2018.
- C.A. Werley, M.P. Chien, J. Gaublomme, K. Shekhar, V. Butty, B.A. Yi, J.M. Kralj, W. Bloxham, L.A. Boyer, A. Regev, A.E. Cohen, Geometry-dependent functional changes in iPSC-derived cardiomyocytes probed by functional imaging and RNA sequencing, PloS one, 12.3, e0172671, 2017.
- H.McNamara, H. Zhang, C.A. Werley, A.E. Cohen, Optically controlled oscillators in an engineered bioelectric tissue, Phys. Rev., X 6, 031001, 2016. Supplementary material.
Membrane and DNA mechanics
The mechanical properties of cellular materials modulate their biochemical function, but mechanical forces are difficult to measure in situ. The lipid bilayer plasma membrane is often thought of as a two-dimensional fluid. We found that in mammalian cells, the presence of transmembrane proteins tethered to the underlying cytoskeleton induces a transition to a gel-like state. As a result, membrane tension can be highly heterogeneous within individual cells. We are studying the mechanics and biology of local changes in membrane tension, and how cells regulate the propagation of these changes.
The mechanical properties of DNA are also important for governing its function. Double-stranded DNA is stiff, but single-stranded DNA is flexible. Under high curvature, double-stranded DNA can melt to become locally single-stranded. We studied the interaction of DNA bending, melting, and underlying sequence.
Membrane mechanics
- A.E. Cohen, Z. Shi Do Cell Membranes Flow Like Honey or Jiggle Like Jello?, BioEssays, 42, 1900142, 2020.
- Z. Shi, Z.T. Graber, T. Baumgart, H.A. Stone, A.E. Cohen, Cell membranes resist flow, Cell, 175, 1-11, 2018.

DNA mechanics
- M.J. Shon, A.E. Cohen, Nano-mechanical measurements of protein-DNA interactions with a silicon nitride pulley, Nucl. Acids Res., 44, e7, 2016.
- A. Fields, E. Meyer, A.E. Cohen, Euler buckling and nonlinear kinking of double-stranded DNA, Nucl. Acids Res., 41, 9881-9890, 2013.